2 Getting started with R
2.0.1 Working in R via RStudio
R comes with its own GUI depicted in Figure 2.1. Nearly everything in R happens either in the script editor (R editor - Figure 2.1, left window) where the user writes code, or in the console (R console - Figure 2.1, right window) which runs commands and prints results.
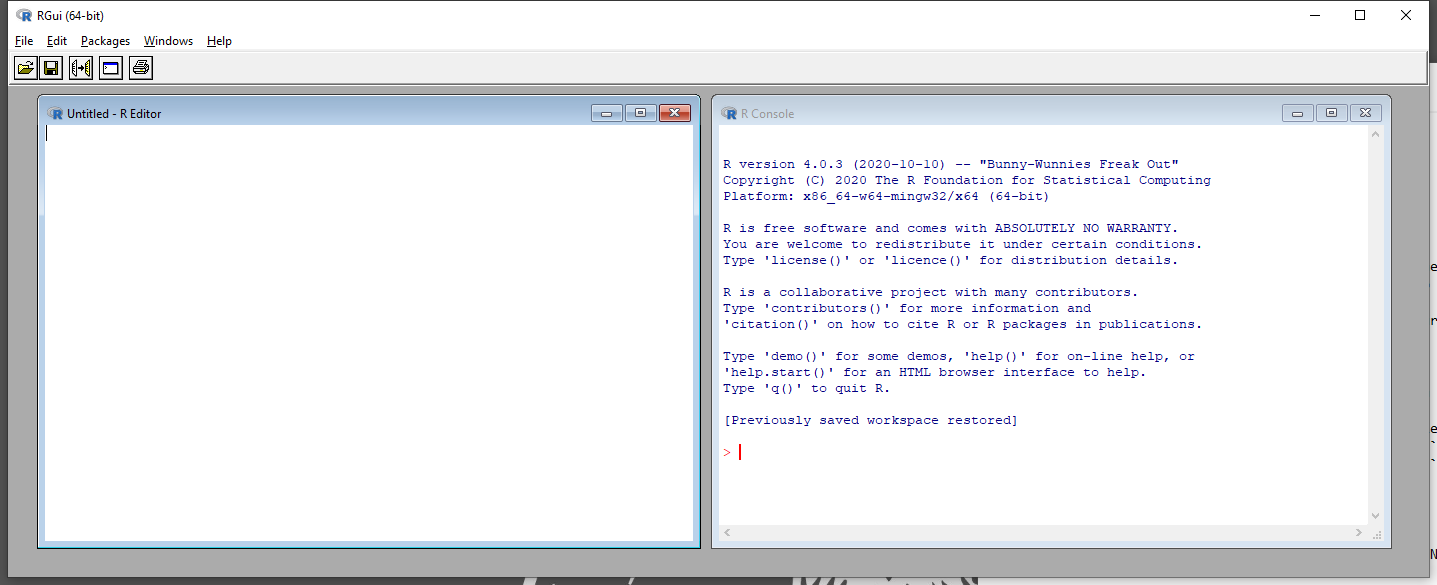
Figure 2.1: R Graphical User Interface.
It is a minimalist GUI that does not offer much more than a short main menu and five buttons in total. It is the engine that will do all your analyses, but does not look very user-friendly.
Fortunately, other programs can fix that. RStudio is one of them.
2.0.2 What is RStudio?
RStudio is an integrated development environment (IDE) for the R language. RStudio runs R “in the background”, and replaces its minimalist interface with its own. This means that you do not lose anything of R’s power, you simply work with it from a different perspective.
Figure 2.2 shows RStudio’s GUI.
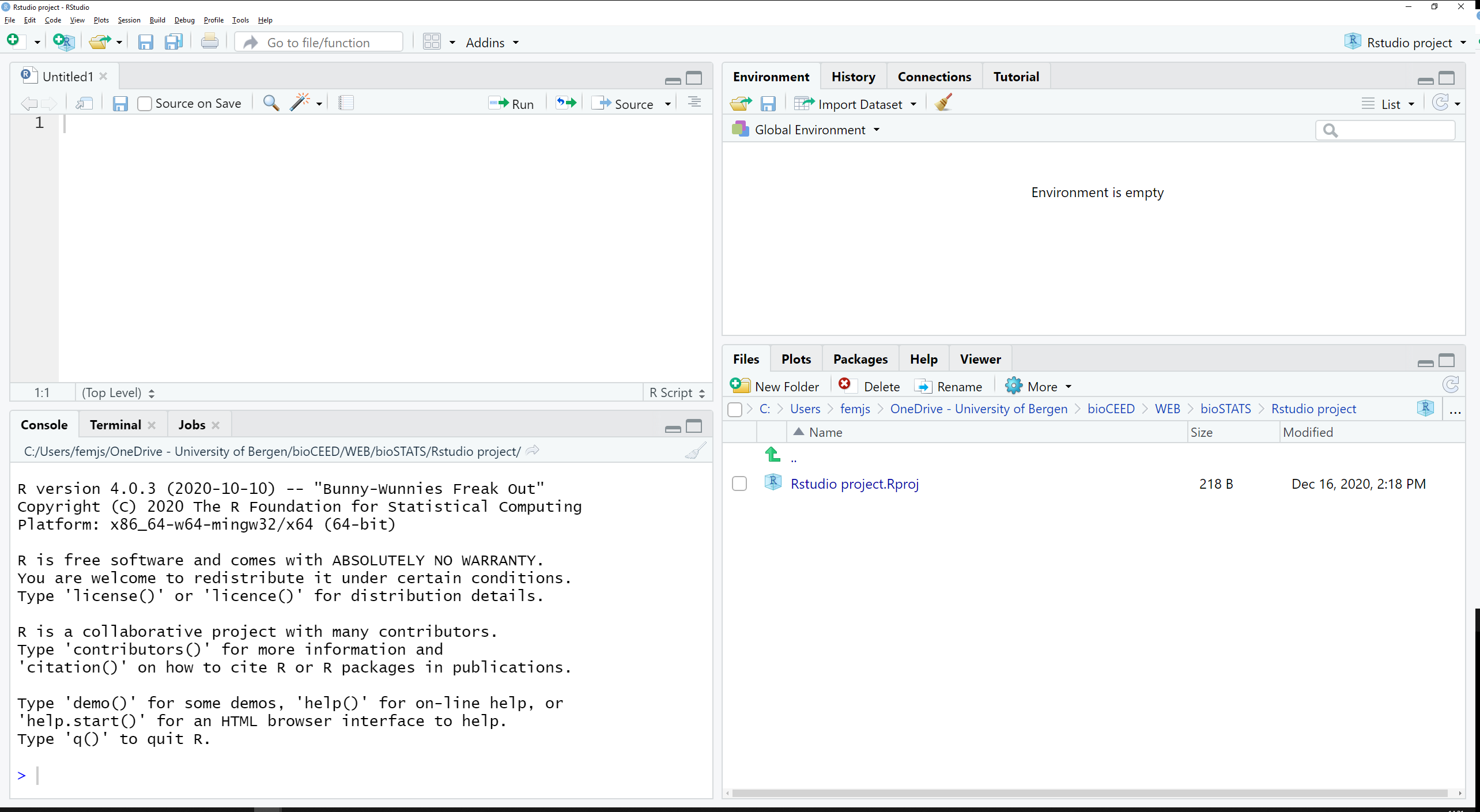
Figure 2.2: RStudio Graphical User Interface.
This interface is more complex and organized.
The script editor (top left) is more advanced and equipped with a syntax highlighter, which will prove useful when writing code.
Many functions (console, file explorer, etc) are available and dispatched in panes and tabs.
A significant benefit of using RStudio is the possibility to create and manage projects.
Projects let you organize your tasks and load only the files and packages that you define as necessary for the workflow.
We will further describe RStudio’s interface in section 2.2.1 and how to set up projects in section 2.4. For now, let’s install everything we need, starting with R and RStudio.
2.1 Installing R and RStudio
There are several ways to install R and RStudio on your machine, depending on whether you are a UiB/NTNU student or staff, or neither. The following sections give you the way to proceed in most cases.
2.1.1 UiB student
UiB students can access and download R and RStudio in a virtual machine, through the Third Party Portal (apps.uib.no). Simply go to apps.uib.no, search for R and RStudio and install both on your virtual machine. You will find information about how to log on and use the Third Party Portal for the first time here in Mitt UiB.
2.1.2 UiB staff
UiB staff working on a client setup machine should use the app “Software Center” (Windows 10) or “Managed Software Center” (Mac OS) to install R and RStudio. You will find help with programme installation here (Windows) and there (Mac OS).
UiB staff who wish to install R and RStudio on a virtual machine can use the Third Party Portal (apps.uib.no). Simply go to apps.uib.no, search for R and RStudio and install both on your machine. You will find information about how to log on and use the Third Party Portal here in Mitt UiB.
2.1.3 NTNU student
NTNU students can access and download many programmes, including R and RStudio, through the software.ntnu.no. Simply go to software.ntnu.no, search for R and RStudio and install both on your machine.
2.1.4 NTNU staff
NTNU staff working on a client setup Windows machine shall use the app “Software Center” that comes preinstalled to install R and RStudio. A description of how to find and use Software Center can be found here. Search for R and RStudio in “Applications”, and install both on your machine.
Alternatively, go to software.ntnu.no, search for R and RStudio and install both.
2.1.5 Anyone
2.1.5.1 Installing R
Go to The Comprehensive R Archive Network. In the top section “Download and Install R”, click on the link that matches your platform and follow the instructions to install the version of R designed for your OS.
2.1.5.2 Install RStudio
Go to RStudio’s website and download the free version of RStudio Desktop made for your OS. Install it on your machine.
Exercise
Install R and RStudio either on your laptop (UiB students can use Third Party Portal instead). You need R version 4.1 or newer and a recent version of RStudio. If you installed R or RStudio more than a few months ago, you will need to reinstall it.
2.2 Starting with RStudio
Now that both R and RStudio are installed on your machine, you are ready to start. Note that you do not need to do anything to link RStudio to R or vice versa. Everything should be ready to use.
2.2.1 The interface
When opening RStudio for the first time, the following main screen (Figure 2.3) appears: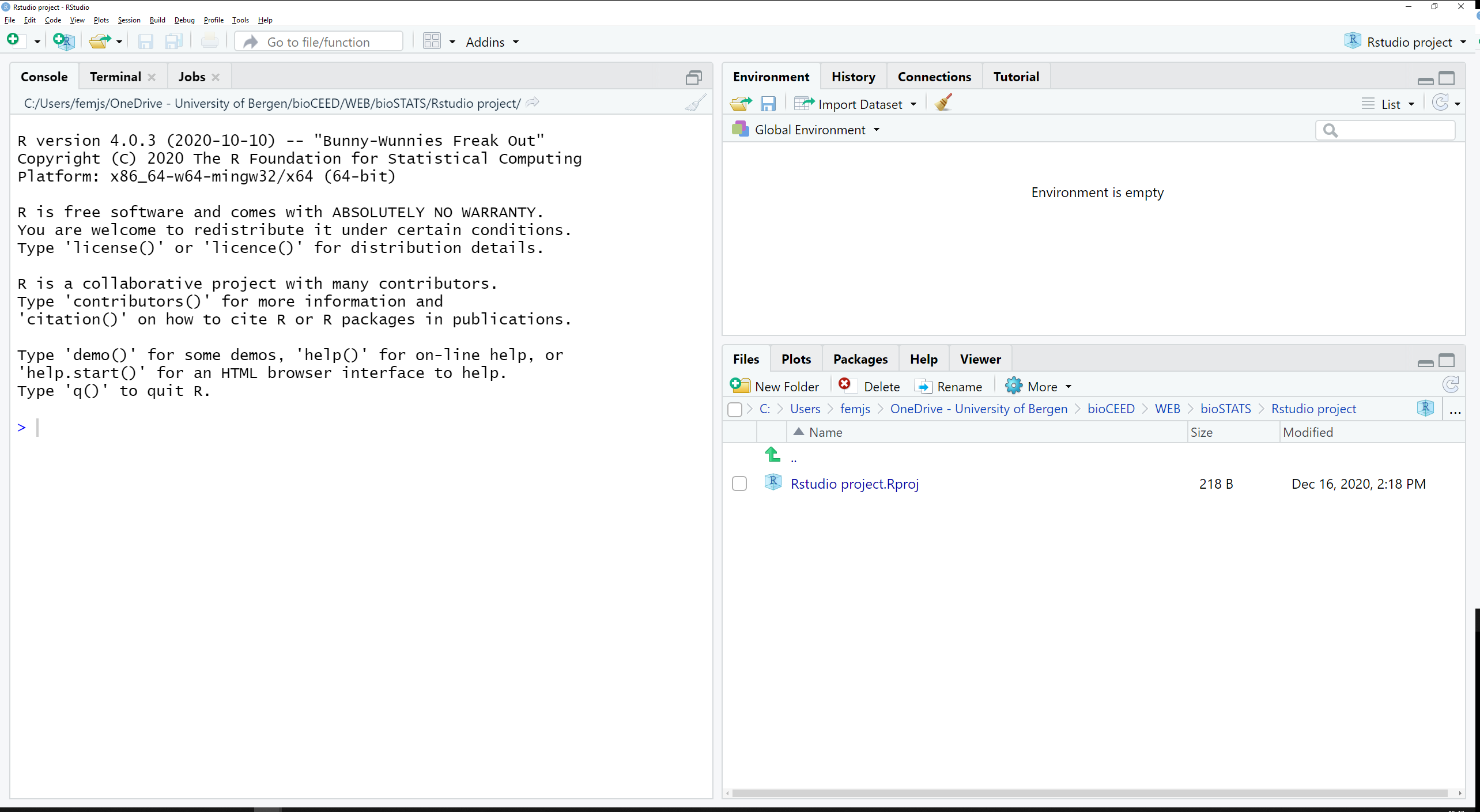
Figure 2.3: Opening RStudio for the first time.
The interface is divided into 3 panes:
- a large one to the left that contains the tabs
Console,TerminalandJobs, - a smaller one in the top right quadrant with the tabs
Environment,History,ConnectionsandTutorial, - a last one in the bottom right quadrant with the tabs
Files,Plots,Packages,HelpandViewer.
In the upcoming sections, we will see what these tabs are made for (NB: only the tabs in bold will be dealt with here).
2.2.2 Scripts
First, go to the main menu in File > New File and choose R Script.
This opens a new, empty tab called Untitled1 in the top left pane (see Figure 2.4).
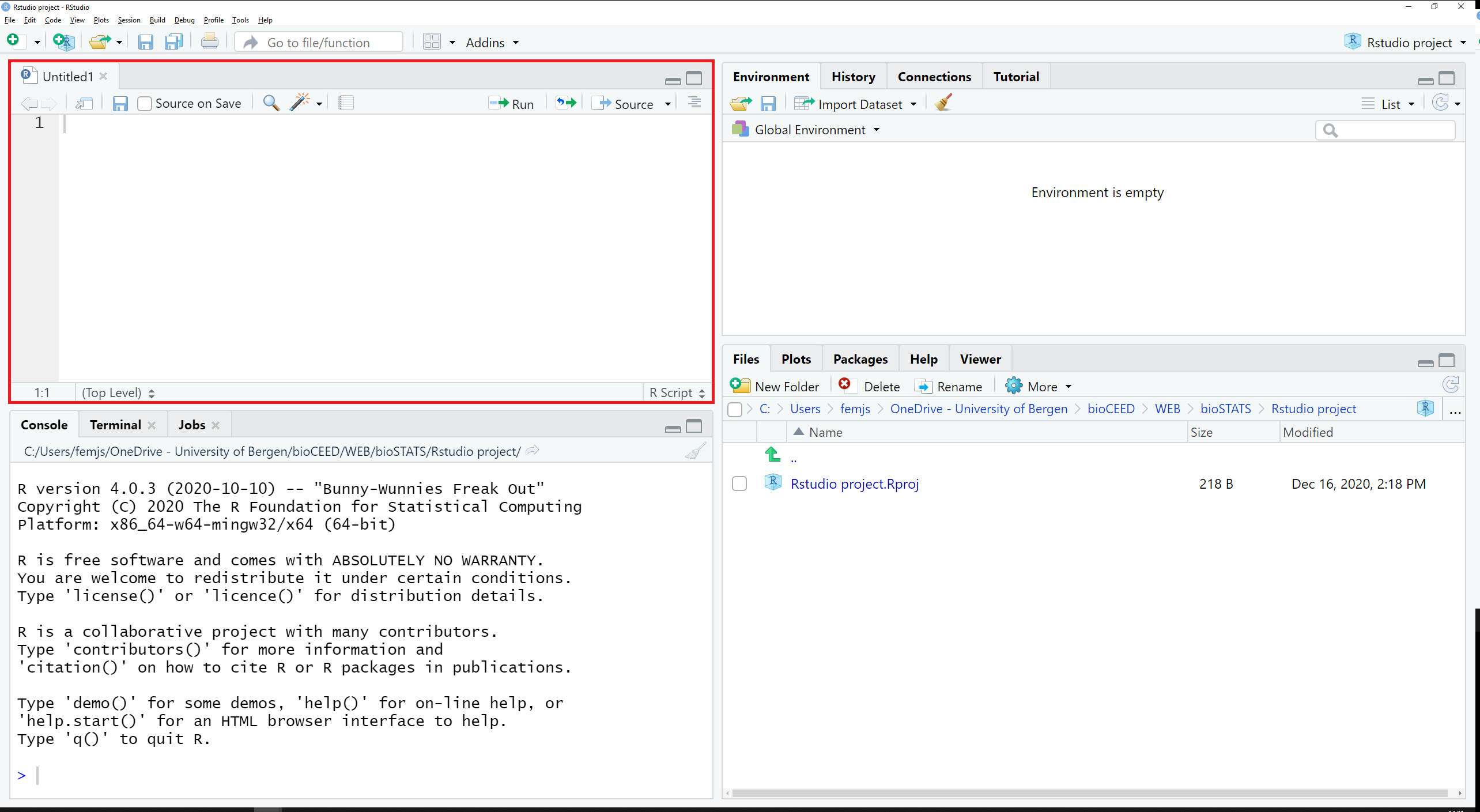
Figure 2.4: The script Untitled1 shows up in the top left pane.
This tab displays a script.
A script is a worksheet that looks a lot like a plain text file.
This is where you will write your code, edit it, correct it if necessary.
It may contain just a few lines, or hundreds of them.
It may also contain comments (lines starting with the symbol #) which will help you keep track of your work.
Here is a simple script:
# this is my data
simple_data <- c(4, 5, 9, 75, 2, 11, 8, 45, 61, 64, 54, 5, 4, 4, 16, 65, 4, 65, 1, 56, 16, 5, 49, 4, 65)
# calculate the mean
mean(simple_data)
# calculate the standard deviation
sd(simple_data)You may open several scripts at the same time.
Each of them will show up as a separate tab in the top left pane of RStudio.
If the tab title is red and followed with a star *, this means that the script is not yet saved, or has been edited since the last time it was saved.
Scripts may be saved at any time using CTRL + S (or ⌘ + S).
In section 2.5, we will talk more about working with scripts and adding comments.
2.2.3 Console and Terminal
The tabs Console and Terminal are located in the bottom left pane of RStudio, along with Jobs (see Figure 2.5).
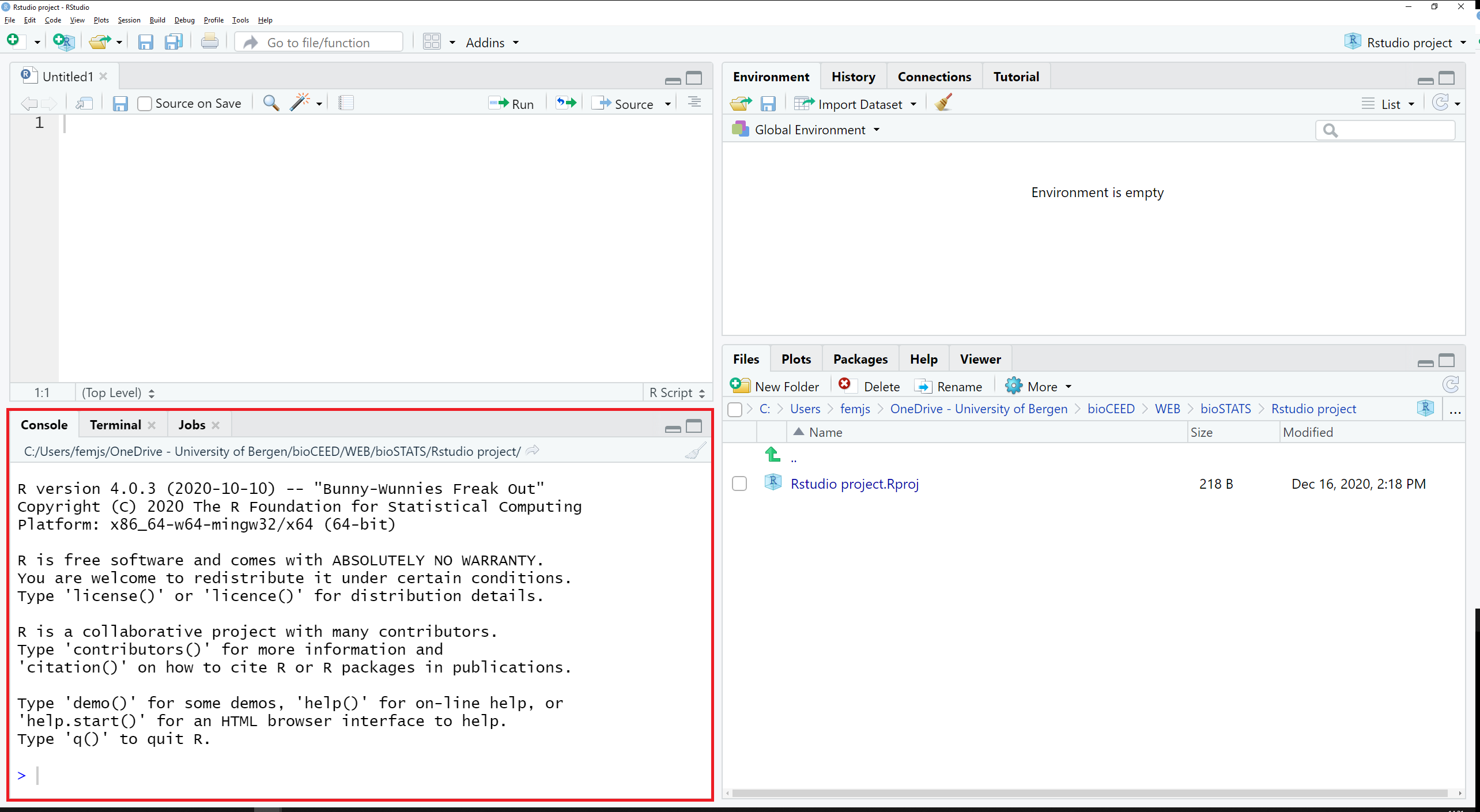
Figure 2.5: The tabs Console and Terminal are located in the bottom left pane.
2.2.3.1 The Console tab
The console is the R module that executes the commands. This is where you find the output/results of your commands providing that they can be display with symbols or characters (as opposed to graphics).
A greater-than sign > displays at the beginning of the line.
This is the prompt.
In the console, every command that you enter at the prompt appears in blue; the output of your commands is printed in black, and errors or warning messages appear in red (see Figure 2.6).
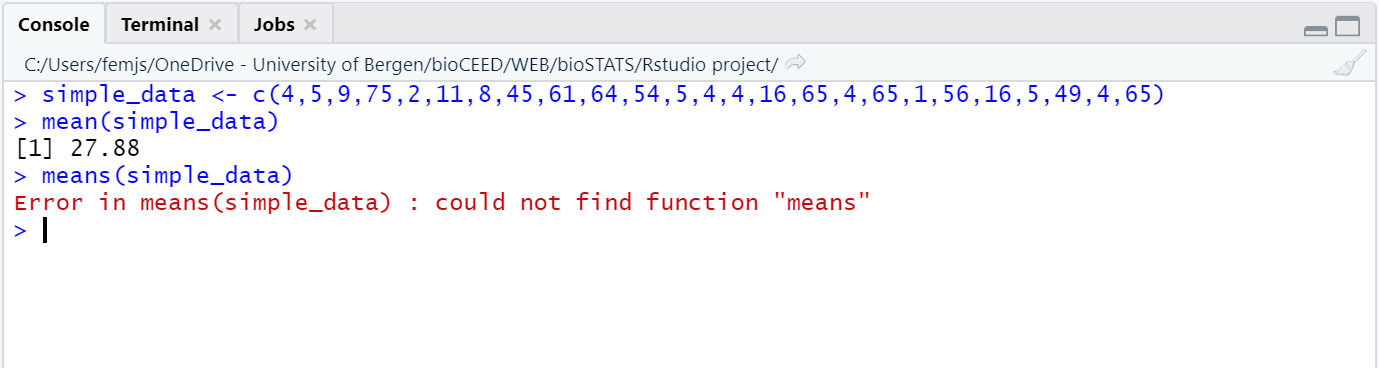
Figure 2.6: The console prints code, output and messages in different colors.
You can actually write a simple command directly in the console and run it with Enter, but this is not good practice: one should always write code in the script and run it in the console (see section 2.5).
The exception is for code that you don’t want to run again, such as code to install a package.
If a plus sign + appears instead of >, that means that your command is incomplete (you are possibly missing a bracket or a quote mark) and R is waiting for something more.
You may either complete the code, or press Esc to return to the prompt.
2.2.3.2 The Terminal tab
The tab Terminal allows for manipulating files locally on your machine or remotely on a server, running Python scripts, etc (see Figure 2.7).
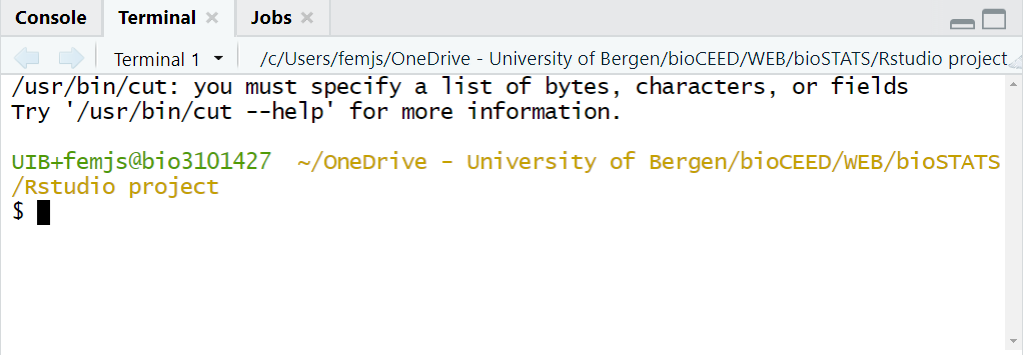
Figure 2.7: The tab Terminal allows for running commands outside of the R environment.
2.2.4 Files, Plots and Packages
The tabs Files, Plots and Packages are located in the bottom right pane, along with Help and Viewer (see Figure 2.8).
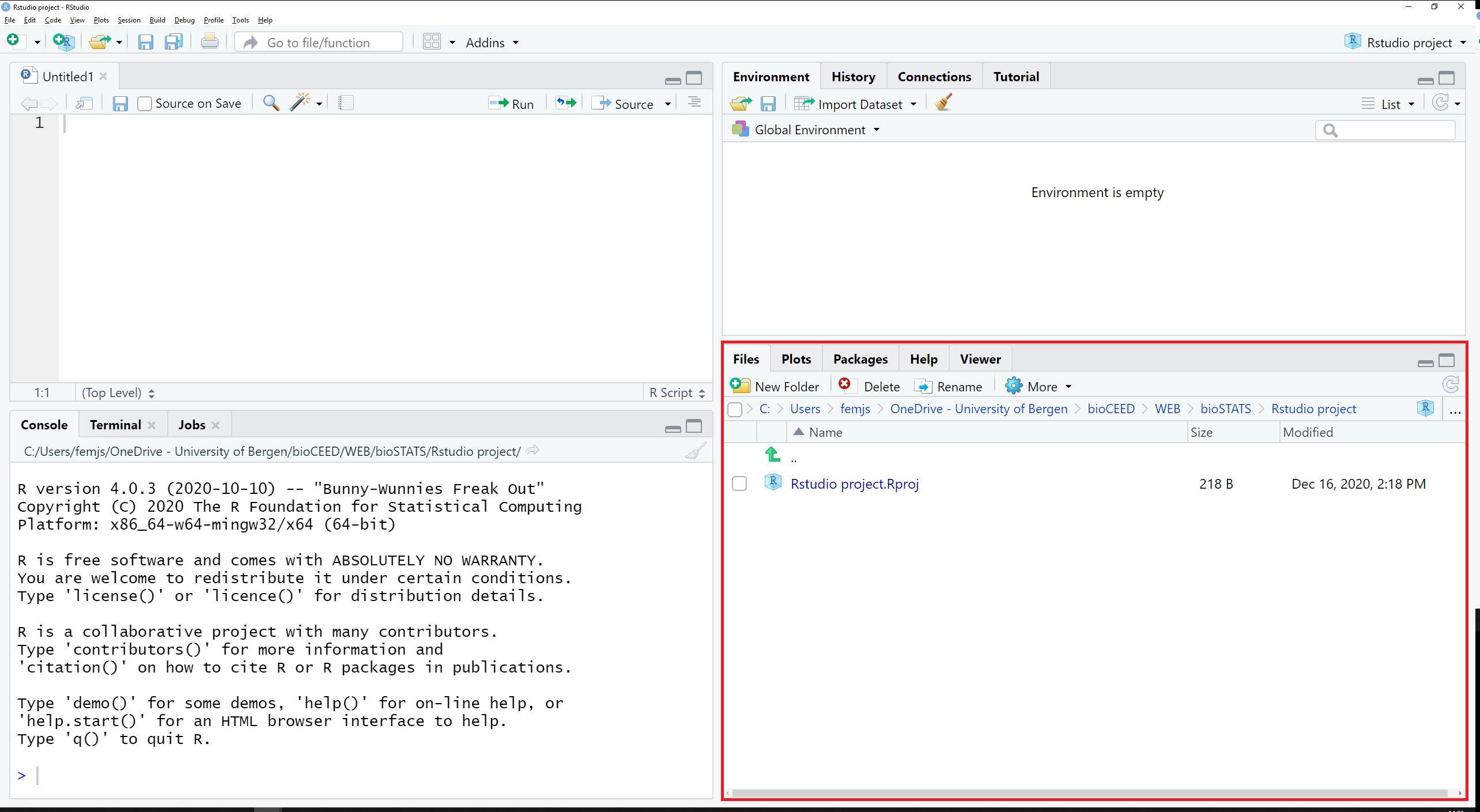
Figure 2.8: The tab Files, Plots and Packages are located in the bottom right pane.
2.2.4.1 The Files tab
The tab Files is a file explorer that lets you navigate the folder structure of your project (for more info about projects, see section 2.4).
When RStudio starts up in a given project, the tab Files displays by default the content of the project folder.
For a new project, the only content should be able to see is a single .Rproj file.
NB: We will see in section 2.4 what the benefits to work with a project are.
This is also the folder where the scripts that you create are preferentially saved and stored.
Feel free to add subfolders, data files and anything else that will be relevant for your work.
Figure 2.9: The menu in the tab Files
Via this menu, you can rename and/or delete the files you have checked in the list beforehand; you can also create new folders, and copy or move items to other places via the dropdown menu of the button More.
2.2.4.2 The Plots tab
The tab Plots is the place where graphic outputs that result from your code will be displayed.
Whenever a code chunk leading to a plot is run in the console, the corresponding plot appears in that tab and its size will adapt automatically to the size of the pane. When changing the dimensions of the pane, plots will be automatically refreshed to fit the new frame.
At the top of the pane, you will find the following menu:
Figure 2.10: The menu in the tab Plots
Via this menu, you can explore all the plots that have been created (not only the latest one) with the arrows, zoom in and out, delete the current plot or all the plots.
The button Export offers two options to save the currently displayed plot as a file.
You may either save as image or save as pdf.
In both cases, a dialog box pops up that lets you define the dimensions, target folder, file name, file type, etc.
2.2.4.3 The Packages tab
The tab Packages provides you with a list of all the R packages that are currently installed on your machine (see Figure 2.11).
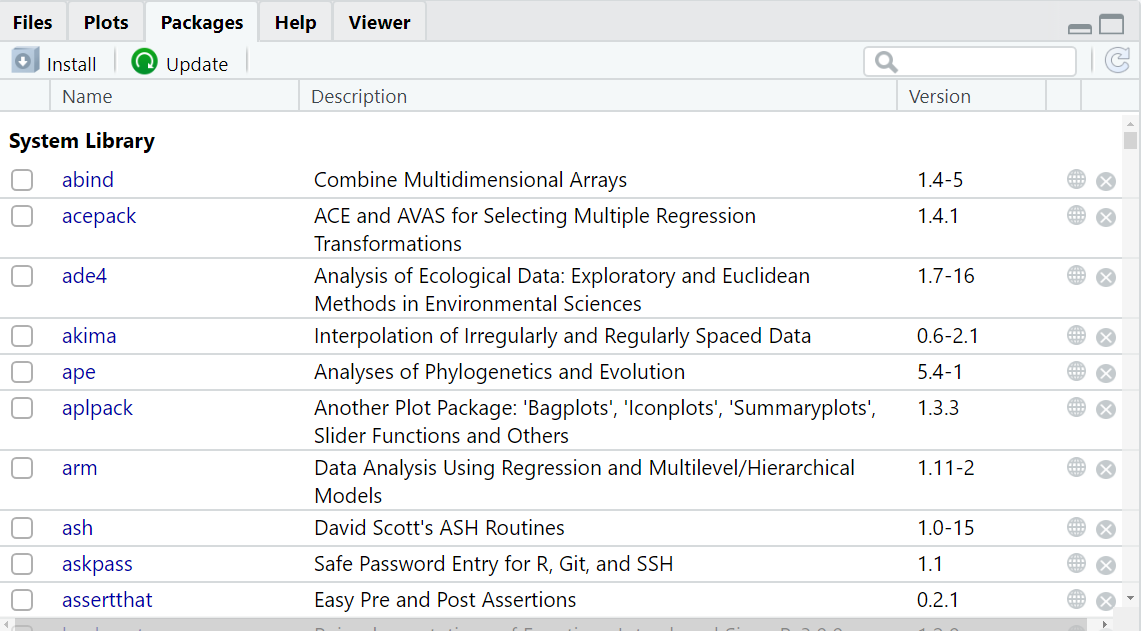
Figure 2.11: The tab Packages shows the packages installed on your machine.
Each line corresponds to a specific package. The checkbox to the left indicates whether the package is currently loaded in RStudio or not, in which case any command referring to it will not perform properly. A short description of the package comes along, as well as the version of the package currently installed. Conveniently, the globe icon to the right brings you to the online information page, and the cross icon allows you to uninstall the package.
Only two items are found in the menu (depicted in Figure 2.12):
-
Install, which also you to install new packages from a remote repository or a file on your machine, -
Update, which searches for newer versions of the packages that are already on your machine.
Figure 2.12: The menu in the tab Packages
2.2.5 Environment and Tutorial
The tabs Environment and Tutorial are located in the top right pane, along with History and Connections (see Figure 2.13).
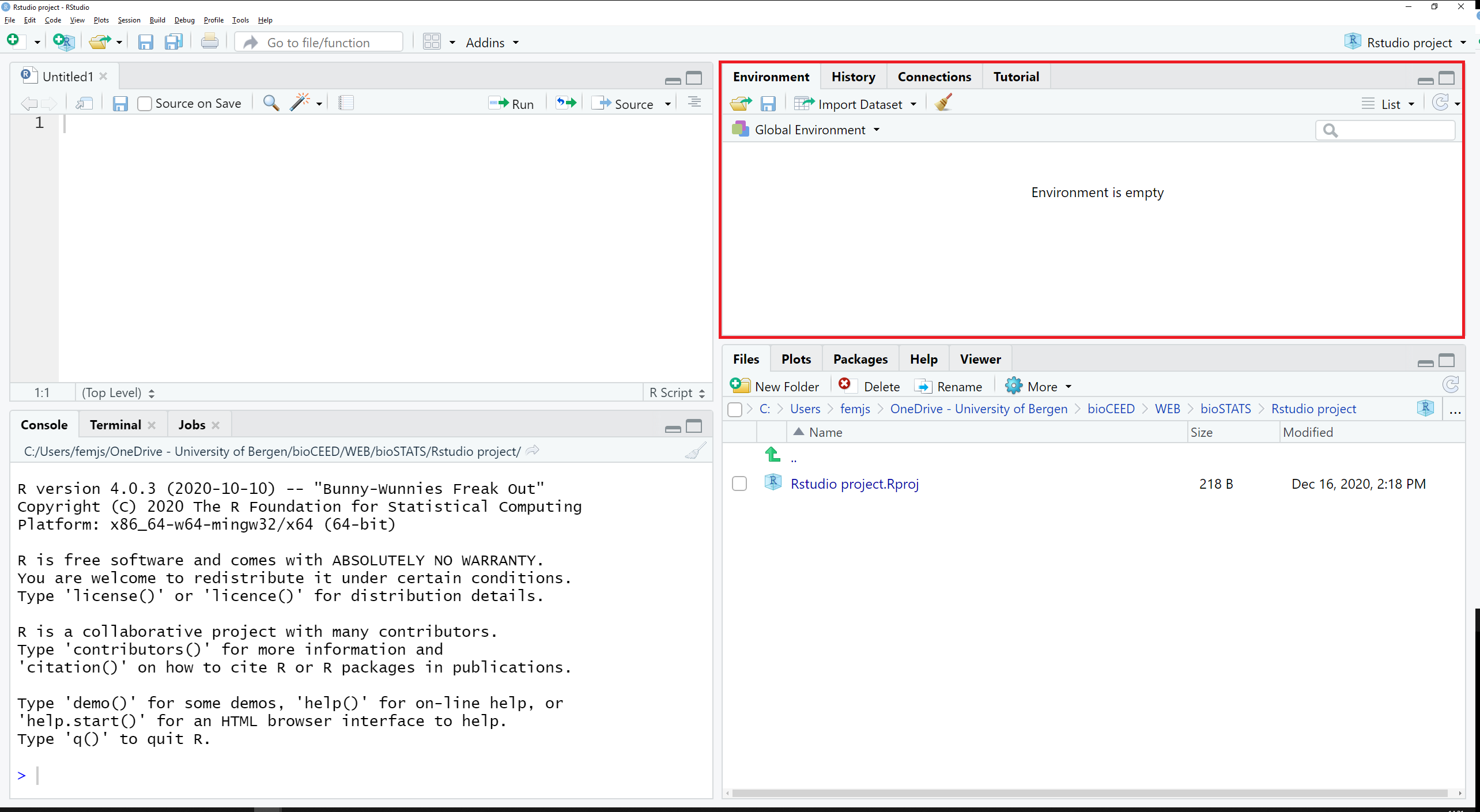
Figure 2.13: The tabs Environment and Tutorial are located in the top right pane.
2.2.5.1 The Environment tab
The tab Environment lists all the R objects currently stored in memory in the current project along with a quick summary of their content.
one_2_three_4, one_two_three, result and results) have been stored in memory.
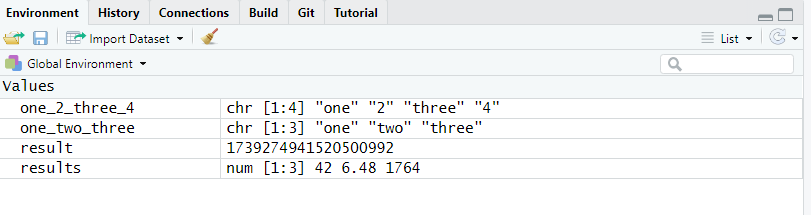
Figure 2.14: The tab Environment shows the R objects stored in memory.
You can see that each object is displayed on its own line, along with a quick overview of its content and nature. You will learn extensively about R objects and data in the 3.
2.2.5.2 The Tutorial tab
The tab Tutorial lists R tutorials which come preinstalled with packages and which may be run directly in this tab (see Figure 2.15).
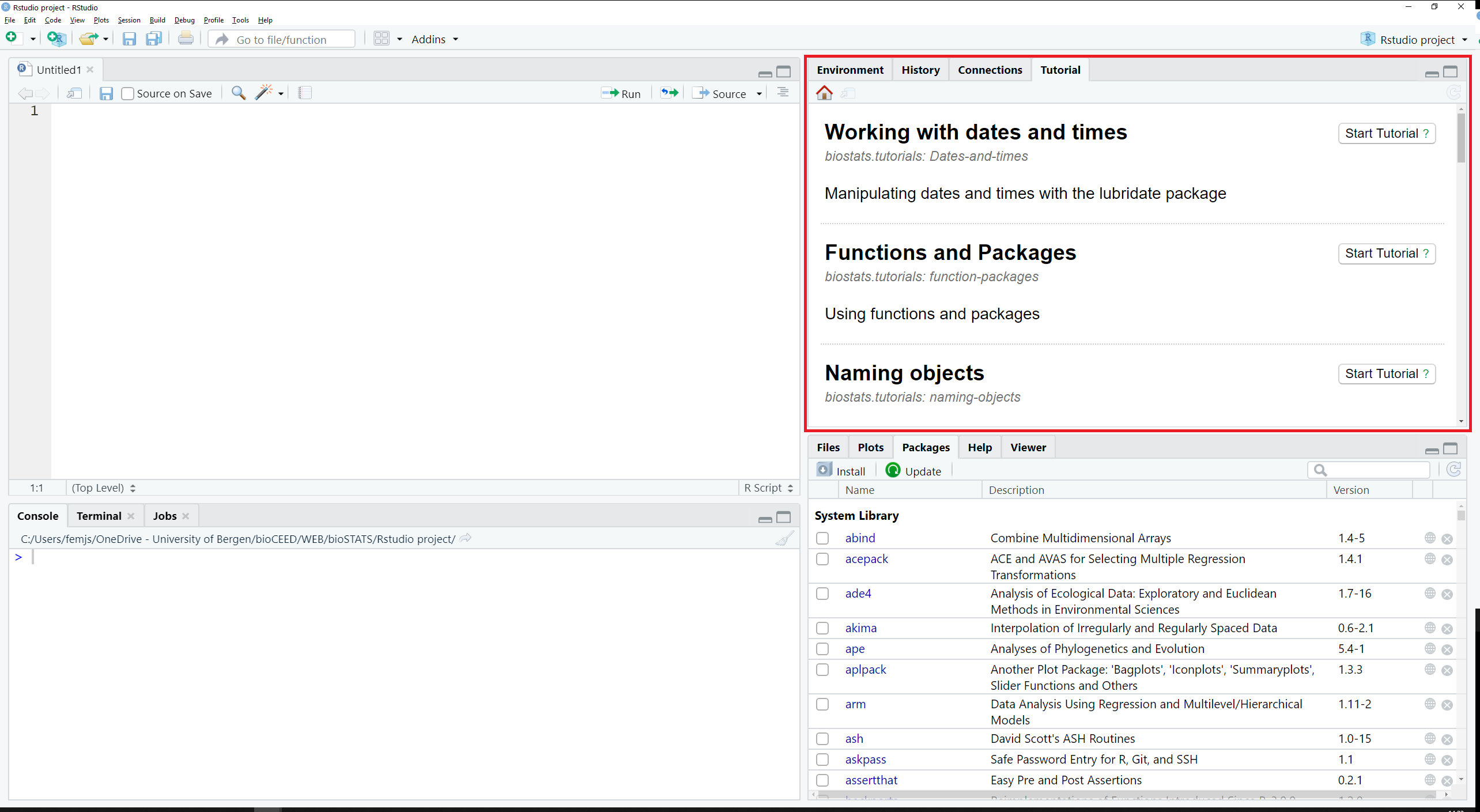
Figure 2.15: The tab Tutorial is located in the top right pane.
Each tutorial is displayed along with a short description, the package it originates from, and a button Start Tutorial ? (see Figure 2.16).
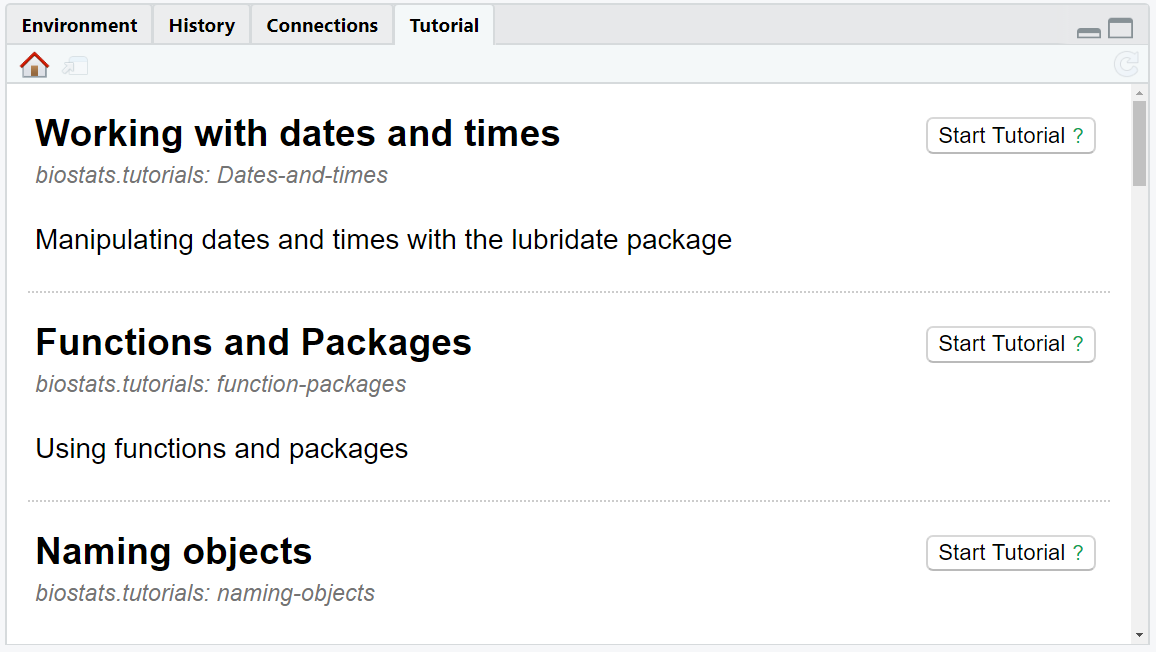
Figure 2.16: The tab Tutorial provides you with tutorials linked to R packages.
Along with the present website, we have written the package biostats.tutorials that will help you better learn stats and R.
The installation procedure is described further below in section 2.3.
Once installed, our tutorials will be available in this tab.
2.3 Installing packages
Packages are add-ons to base R (the R base package) that expand the computing possibilities of R by adding new functions, classes, documentation, data sets, etc.
When installing RStudio for the first time, a long list of packages comes along.
You will find this list in the tab Packages (see section 2.2.4.3).
This section will show you how to install additional packages.
Every time you install a new one, R imports all necessary files into a local library, but does not activate it.
You will have to remember to activate the new package with library() every time your project require items or functions from that package.
2.3.1 Packages published on CRAN
If you want to install a package published on CRAN repository, you may use the function install.packages().
Simply type its name and add quotation marks " ".
Here is an example with the package tidyr:
install.packages("tidyr")You may use the exact same code to update the package later on.
If you want to update all packages at once, use the following code:
2.3.2 Packages published on GitHub
If you want to install a package published on GitHub, you may use the function remotes::install_github().
First you need to install the package remotes.
Type first the name of the repository, then \ and the name of the package.
Here also, you must add quotation marks " ".
2.3.3 Recommended packages
The present website refers to the use of several packages that we recommend, not only for learning R, but also for working with your course assignments and projects. These packages are:
tidyverse-
biostats.tutorials, -
palmerpenguins.
These packages will be used throughout the website, so get ready to install them now. Here is the procedure.
2.3.3.1 The tidyverse
The tidyverse is an amazing toolbox which contains a growing, evolving collection of R packages for data science. The packages are developed on the same philosophy and are fully compatible with each other. Some of the tidyverse packages help you read files and import data, some packages let you draw plots and make figures, others help you rearrange your data set. Have a look at the tidyverse webpages to explore the collection.
Install and activate the tidyverse with the following lines:
install.packages("tidyverse")
library(tidyverse)2.3.3.2 palmerpenguins
palmerpenguins is a package that provides you with two real data sets.
They contain measurements from three penguin species found in the Palmer archipelago, Antarctica.
Several variables such as species, island, and body mass are included, and no less than 344 observations are found in the table.
These data sets will be used in the upcoming sections of this website.
Install and activate the package with these lines:
install.packages("palmerpenguins")
library(palmerpenguins)Type this following line in the console if you want to know more about palmerpenguins:
citation("palmerpenguins")2.3.3.3 biostats.tutorials
Along with the present website, our team has developed a package with tutorials called biostats.tutorials.
This package will help you learn and practice R functions and concepts.
Several of the upcoming chapters refer to this package.
Install biostats.tutorials with this code:
remotes::install_github("biostats-r/biostats.tutorials")Now load the package with library(biostats.tutorials) and the tutorials should appear in the tab Tutorial (see section 2.2.5.2).
Exercises will tell you when to run a tutorial.
Click the “Start Tutorial” button to start the tutorial (this may take a few seconds to start the first time).
Exercise
Install the tidyverse, palmerpenguins from CRAN and biostats.tutorials from GitHub using the code above.
If your R version is < 4.1, follow these instructions
Using R < 4.1?
Ideally upgrade R to the latest version.
If you cannot, run this code to modify biostats.tutorials so that it can run on your computer.
library(tidyverse)
old_pipes <- function(){
if(R.version >= 4.1){return()}
#find Rmd files in tutorial directory
list.files(system.file("tutorials",
package = "biostats.tutorials"),
recursive = TRUE,
pattern = "\\.Rmd",
full.names = TRUE) %>%
#iterate over files and read them
map(~{read_lines(.x) %>%
#replace new pipes with old
str_replace_all(pattern = fixed("|>"), replacement = "%>%") %>%
#write file
write_lines(file = .x)})
}
old_pipes()2.4 Working with a project
RStudio allows you to divide your work into projects which are independent from each other. A project has its on working directory in which you can create specific scripts, load data sets, add external files, activate packages, etc. For each project, you will thus be guaranteed to work in a dedicated workspace.
Working with projects is a great way to keep things tidy. You do not risk to mix up files or variables with relatively similar names when writing your code, especially if you are not so creative when it comes to naming objects. You will also find it easy to share your work with others since everything they need (and nothing less or more) is in a project.
For example, if you plan to work with assignments in different courses in addition to the data analyses for your master’s thesis, we strongly recommend to set up a RStudio project for each course, and another for your thesis.
To create a project, go to the main menu and select File > New Project… as shown in Figure 2.17: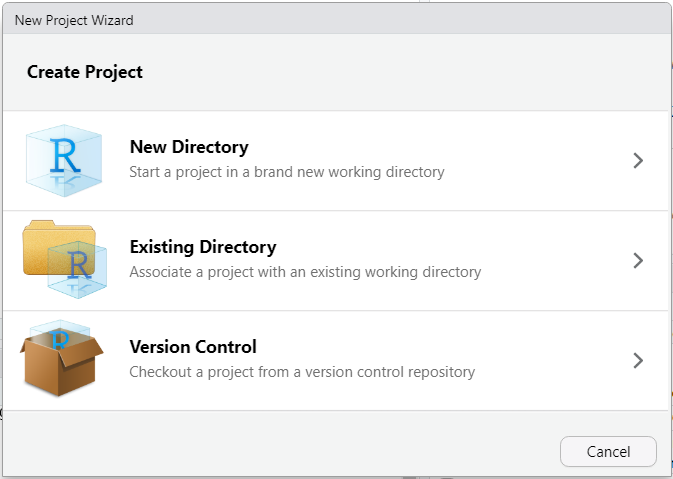
Figure 2.17: Creating a new project – step 1
Click again on New Directory > New Project, choose a project name and a destination on your disk, and click the button “Create Project” as shown in Figure 2.18:
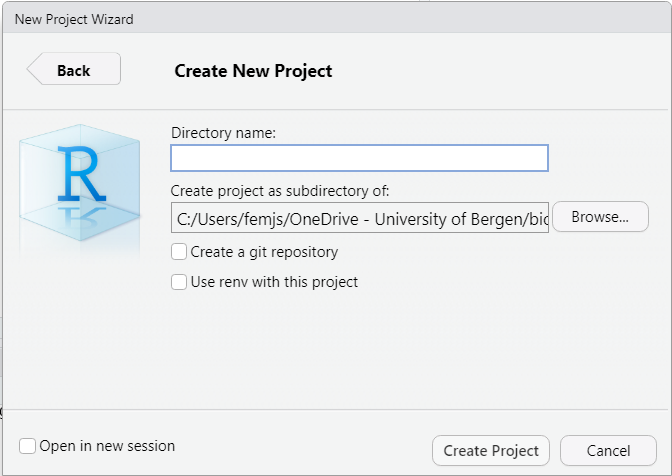
Figure 2.18: Creating a new project – step 2
Feel free to import in the project folder all the files that you will need later on, such as original data sets, etc.
2.5 Working with a script
2.5.1 What is a script?
Practically, a script is a plain text file where you write your code, whether it contains a handful of lines or dozens of them. It is an evolving document which not only helps you keep track of your code, but also your workflow.
With time, you will realize that a script is a lot of things at the same time:
- it is a whiteboard where you try coding for something and correct mistakes whenever you find out things do not work as expected,
- it contains your coding history, where all the steps from loading a data set to printing the final output are chronologically exposed,
- it is the key file that you may share with collaborators, etc,
- it is guarantee that your work is reproducible, meaning that you can run your code on your data set again and again, and obtain the same result, consistently.
A simple script may look like this:
# activate tidyverse
library(tidyverse)
# load the data from external file
Veronica_Vestland <- read_delim("Veronica_Vestland.csv", delim = ",")
# calculate the mean and standard deviation of Sepal.Length for each Location
mean_sd_SL <- Veronica_Vestland |>
group_by(location) |>
summarise(mean(Sepal.Length), sd(Sepal.Length))
# print the result
mean_sd_SL
# draw boxplot Sepal.Length for each Location
ggplot(Veronica_Vestland, aes(x = Location, y = Sepal.Length, fill = Location)) +
geom_boxplot()You will be able to write similar code to this very soon.
2.5.2 Running the code
Writing code in a script does not do anything per se. To tell R to do something, you must either:
- place the cursor on a single line of code and press the
Runbutton above the script or type CTRL + Enter (⌘ + Enter) to run that single line of code, or - select several lines of code and press CTRL + Enter (⌘ + Enter) to run ALL the selected lines at once, or
- press CTRL + ALT + Enter (⌘ + ⌥ + Enter) to run the whole script.
The result of your command(s) will appear in the tab Console if the commands are intended to print something, and/or in the tab Plots if the commands generate a plot.
2.5.3 Code versus comment
In the script above, there are two types of lines: those that start with the symbol #, and those that do not.
Let’s start with the lines that do not start with a #.
They are the real code, the commands that manipulate the data.
Right now these lines do not mean much to you, but in fact, each of them commands R to “do something specific” with your data.
That “something specific” is defined by functions which are followed by parentheses – function().
In the R language, functions are verbs in your sentences, the data are their subject.
For example, in the code above, library(tidyverse) commands R to activate the package tidyverse found in the package library.
The lines that start with a # are comments.
They do not code for anything at all.
When you run the script via a console, R simply ignores them.
So use comments to keep track of what you do with the code.
Write what the point of each real code line is, what you plan to do.
That way, you will always remember what you originally intended to code for, in case you lose track.
The symbol # is also convenient to prevent R from running a specific code line or chunk, without having to delete that line.
Indeed if you place a # in front of any line, the console will consider it as a comment, and simply skip it.
In the following example, each line was originally written to activate a different package:
However, the third line has been commented with a #.
Consequently, only the packages ggplot2, tidyr, and vroom are activated; tidyverse will be ignored.
In the chapter First Steps in R (3), you will learn to write in the R language. We strongly advise you to work in scripts, and make extensive use of comments from the start. This is considered good coding practice, and will save you quite some time and energy.
2.6 Customizing RStudio
R/RStudio does not require much configuring at start, even though the menus in Tools > Global Options… let you change dozens of settings at any time. In fact, you should be ready to work right now. That said, there are a couple of things in RStudio that we recommend you customize.
2.6.1 Taking care of .Rdata
.Rdata is a file that R uses to store objects, data, etc. It is saved automatically when RStudio shuts down, and restored when it starts up again. We advise you to prevent RStudio from saving changes and restoring .Rdata to improve reproducibility.
Go to Tools > Global Options… > General > Basic to get to the menu shown in Figure 2.19. In the section Workspace (Fig. 2.19, red box), uncheck the box, and select “Never” in the dropdown menu.
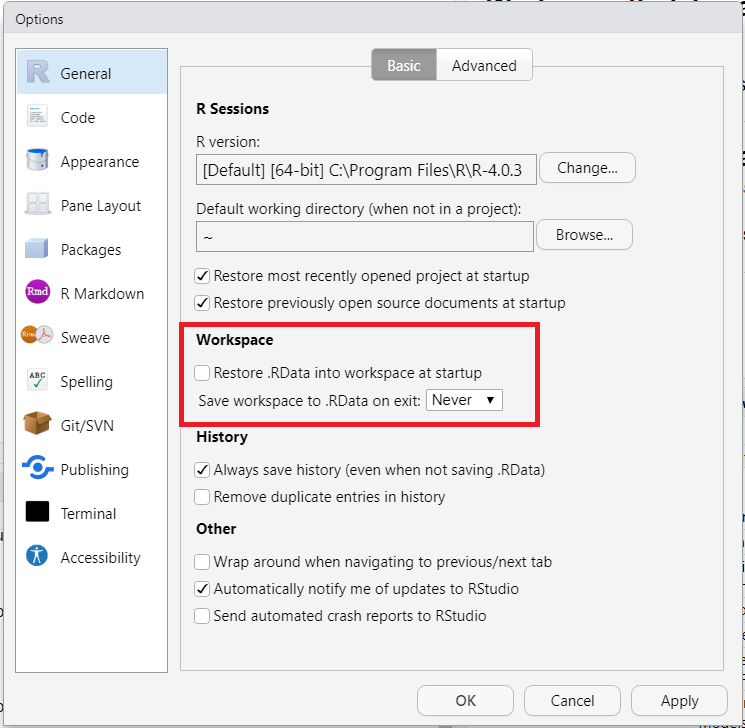
Figure 2.19: Taking care of .Rdata
Or you can do this in code with
usethis::use_blank_slate()2.6.2 Imposing UTF-8 encoding
Not all symbols or characters are accepted or recognized in the format that RStudio uses by default when saving scripts. We recommend to force RStudio to save scripts in UTF-8 format, which is much more permissive.
Go to Tools > Global Options… > Code > Saving to get to the menu shown in Figure 2.20. In the section Serialization (Fig. 2.20, red box), click “Change” and choose UTF-8.
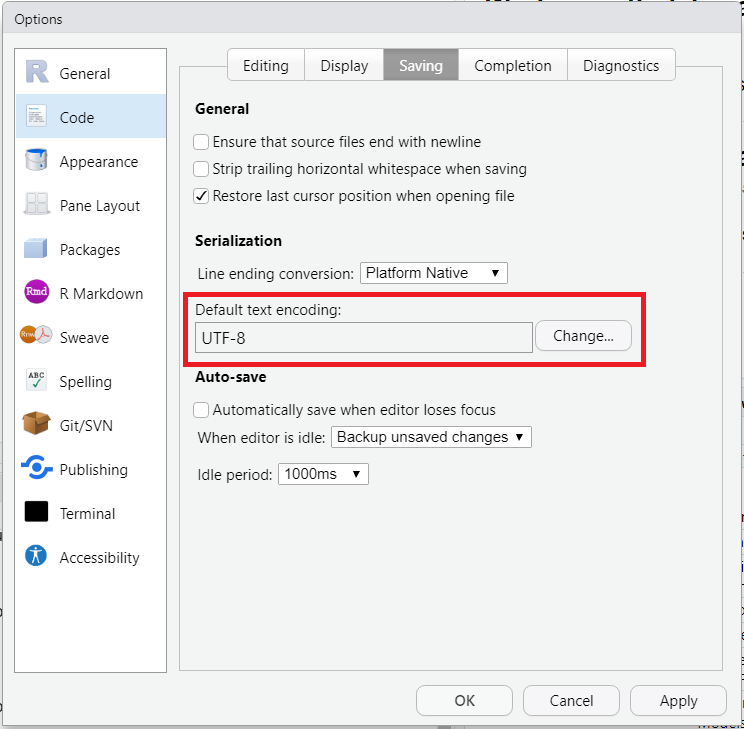
Figure 2.20: Saving files as UTF-8
2.6.3 Soft-wrapping R scripts
When the length of a code line exceeds the width of the editor, a horizontal scrollbar appears at the bottom of the editor, allowing you to navigate and review the whole line from its first to its last character. This setting makes things impractical as you will often have to scroll back and forth when reviewing multiple long lines. The obvious solution is to make sure you write short lines of code - a maximum of 80 characters is often recommended. However on a laptop, this can still be too long, so an alternative is to force RStudio to split the code onto the next line(s) of the editor – this is called soft-wrapping. We recommend that you activate soft-wrapping in RStudio.
Go to Tools > Global Options… > choose Code > Editing to get to the menu shown in Figure 2.21. In the section General (Fig. 2.21, red box), check the box “Soft-wrap R source files”.
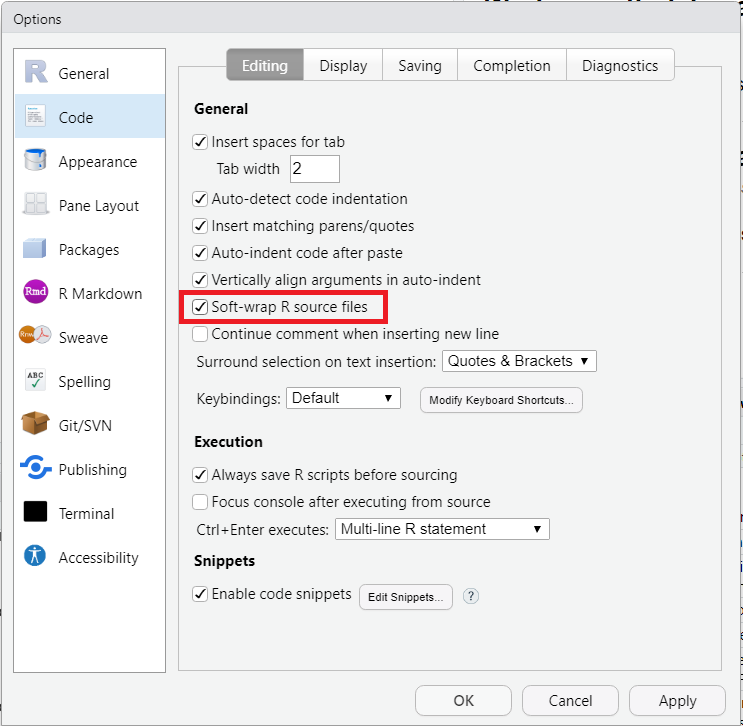
Figure 2.21: Activating soft-wrapping of R scripts
2.7 RStudio keyboard short-cuts
You have already been introduced to RStudio short-cuts to run code. There are many more - press Shift + Alt + k. You certainly don’t need to learn them all. Here are some we find useful
- run line Ctrl + Enter
- Find/replace Ctrl + F
- Find in files Ctrl + Shift + F
- Insert assignment operator Alt + -
- Comment out selected lines Ctrl + Shift + C
- Help on selected function F1
You will be introduced to more short-cuts later.